What is INSIGHT?
INSIGHT is a method for inferring signatures of recent natural selection from patterns of polymorphism and divergence
across a collection of short dispersed genomic elements.
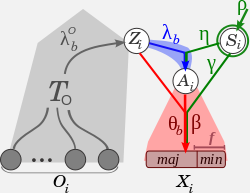
The core of the method is a full probabilistic model that allows it to make use of sequence data across
a dispersed set of elements to disentangle the effects of neutral genetic drift from positive,
strong negative, and weak negative selection.
INSIGHT provides estimates of three main measures:
the fraction of nucleotides under selection (ρ),
the number of divergences driven by positive selection (Dp),
and the number of polymorphisms under weak negative selection (Pw).
Maximum likelihood inference is achieved through an EM algorithm (INSIGHT-EM) implemented in C.
|

|
Download INSIGHT-EM
[ Version 1.1,
last updated on Mar 05, 2013 ]
|
More info
-
INSIGHT was developed by Ilan Gronau and Leonardo Arbiza at the Siepel computational
genomics lab in Cornell University, with contributions from Melissa Hubisz and Jaaved Mohammed.
-
Cite: Gronau I, Arbiza L, Mohammed J, Siepel A.
Inference of Natural Selection from Interspersed Genomic
Elements Based on Polymorphism and Divergence.
Mol Biol Evol, 30(5):1159–1171, 2013.
-
Additional references:
* Arbiza L, Gronau I, Aksoy BA, Hubisz MJ, Gulko B, Keinan A, Siepel A.
Genome-wide inference of natural selection on human transcription factor binding sites.
Nature Genetics, 45:723-729, 2013
* Mohammed J, Bortolamiol-Becet D, Flynt A, Gronau I, Siepel A, Lai E.
Adaptive evolution of testis-specific, recently-evolved, clustered miRNAs in Drosophila.
RNA, in review, 2014
-
For questions and suggestions contact

|



