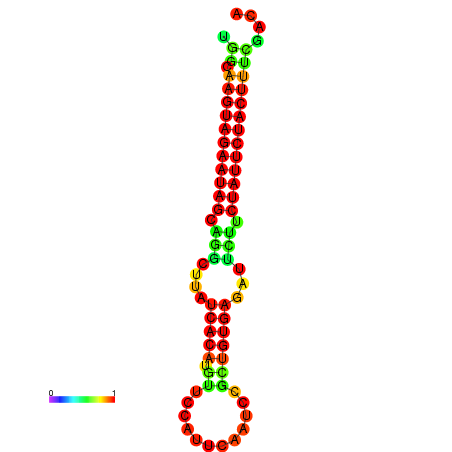
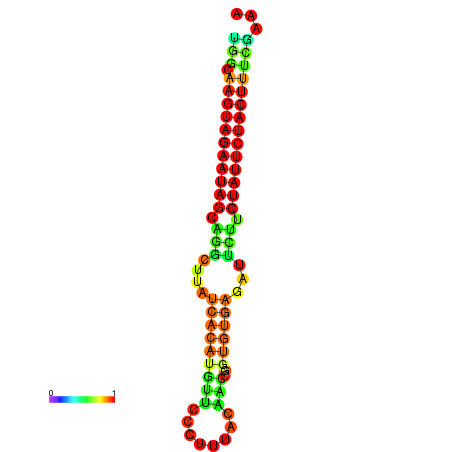
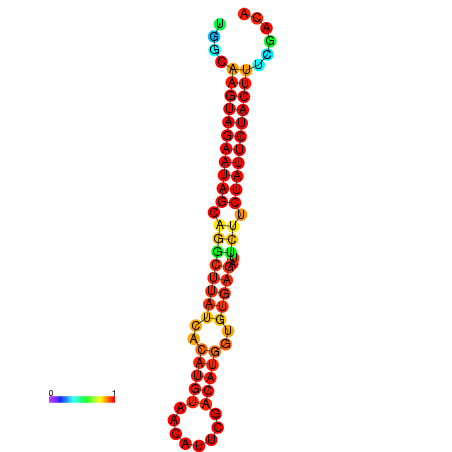
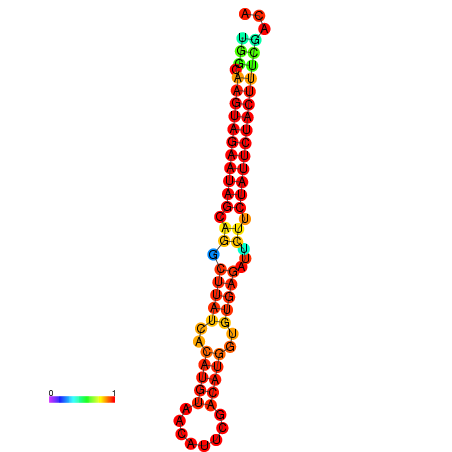
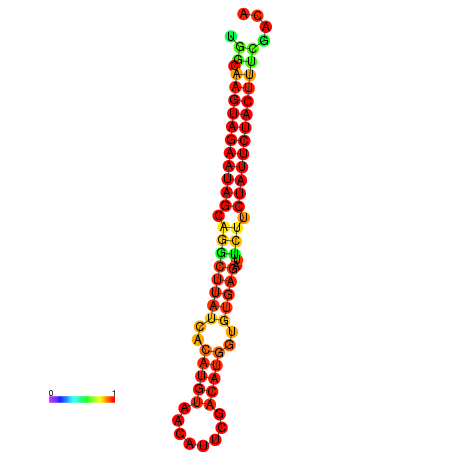
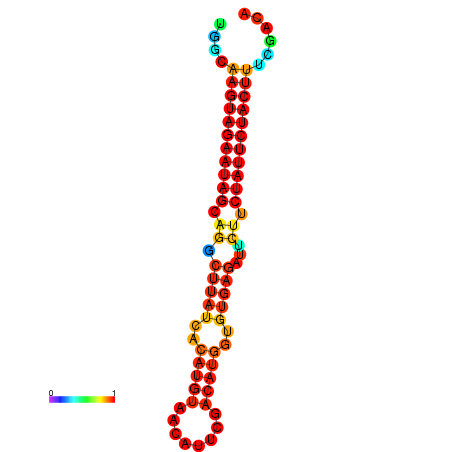
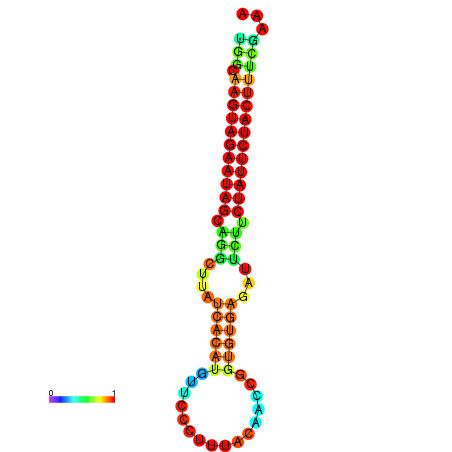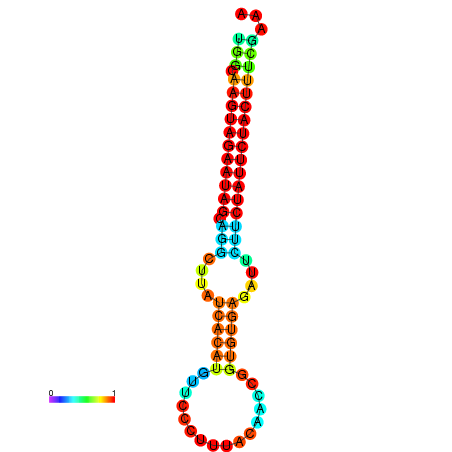| GGATATGGCTAGCACGATCGGTGTGTATTTGGCAAGTAGAATAGCAGGCTTATCACATGTTCCATTCAATCCGCTGTGAGATTCTTCTATTCTACTTTCGACAAACACCCGTTGACATCTAATAGAGCAGCCCTC | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| ..................................AGTAGAATAGCAGGCTTATCACA.............................................................................. | 23 | 1 | 1985.00 | 1985 | 1926 | 59 |
| ............................................................................TGAGATTCTTCTATTCTACTTT..................................... | 22 | 1 | 1302.00 | 1302 | 1099 | 203 |
| ..................................AGTAGAATAGCAGGCTTATCAC............................................................................... | 22 | 1 | 440.00 | 440 | 434 | 6 |
| .........................................................TGTTCCATTCAATCCGCTG........................................................... | 19 | 1 | 76.00 | 76 | 75 | 1 |
| ............................................................................TGAGATTCTTCTATTCTACTT...................................... | 21 | 1 | 69.00 | 69 | 45 | 24 |
| ..................................AGTAGAATAGCAGGCTTATCA................................................................................ | 21 | 1 | 60.00 | 60 | 40 | 20 |
| ..................................AGTAGAATAGCAGGCTTATC................................................................................. | 20 | 1 | 31.00 | 31 | 28 | 3 |
| ....................................TAGAATAGCAGGCTTATCACA.............................................................................. | 21 | 1 | 29.00 | 29 | 29 | 0 |
| ............................................................................TGAGATTCTTCTATTCTACT....................................... | 20 | 1 | 19.00 | 19 | 11 | 8 |
| ....................................TAGAATAGCAGGCTTATCAC............................................................................... | 20 | 1 | 8.00 | 8 | 8 | 0 |
| ..................................AGTAGAATAGCAGGCTTAT.................................................................................. | 19 | 1 | 8.00 | 8 | 8 | 0 |
| .............................................................................GAGATTCTTCTATTCTACTTT..................................... | 21 | 1 | 6.00 | 6 | 6 | 0 |
| ....................................TAGAATAGCAGGCTTATCACATG............................................................................ | 23 | 1 | 4.00 | 4 | 4 | 0 |
| ............................................................................TGAGATTCTTCTATTCTAC........................................ | 19 | 1 | 4.00 | 4 | 4 | 0 |
| ..................................AGTAGAATAGCAGGCTTA................................................................................... | 18 | 1 | 3.00 | 3 | 3 | 0 |
| ............................................................................TGAGATTCTTCTATTCTACTTTC.................................... | 23 | 1 | 3.00 | 3 | 3 | 0 |
| ................................................................................ATTCTTCTATTCTACTTT..................................... | 18 | 1 | 1.00 | 1 | 1 | 0 |
| .....................................AGAATAGCAGGCTTATCACA.............................................................................. | 20 | 1 | 1.00 | 1 | 1 | 0 |
| ......................................GAATAGCAGGCTTATCACA.............................................................................. | 19 | 1 | 1.00 | 1 | 1 | 0 |
| .......................................AATAGCAGGCTTATCACA.............................................................................. | 18 | 1 | 1.00 | 1 | 1 | 0 |
| ............................................................................TGAGATTCTTCTATTCTA......................................... | 18 | 1 | 1.00 | 1 | 1 | 0 |
| ....................................TAGAATAGCAGGCTTATCACATGT........................................................................... | 24 | 1 | 1.00 | 1 | 1 | 0 |
| .....................................AGAATAGCAGGCTTATCAC............................................................................... | 19 | 1 | 1.00 | 1 | 1 | 0 |
| ...................................GTAGAATAGCAGGCTTATCACA.............................................................................. | 22 | 1 | 1.00 | 1 | 1 | 0 |