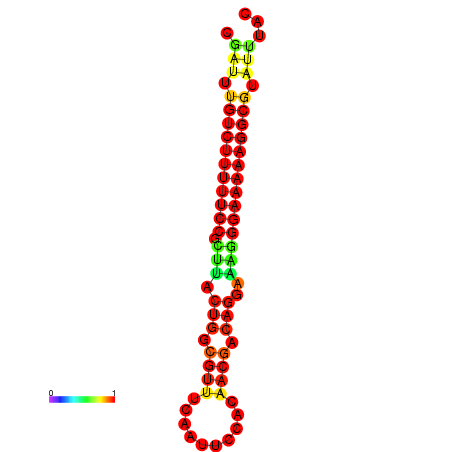
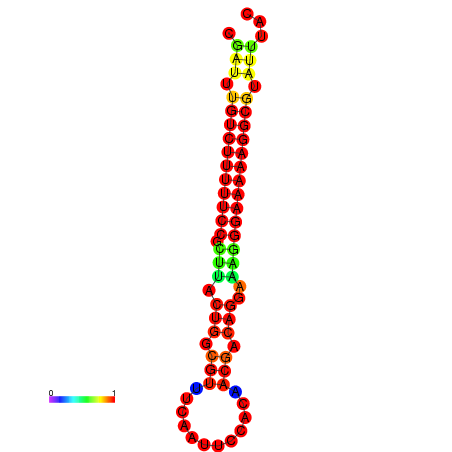
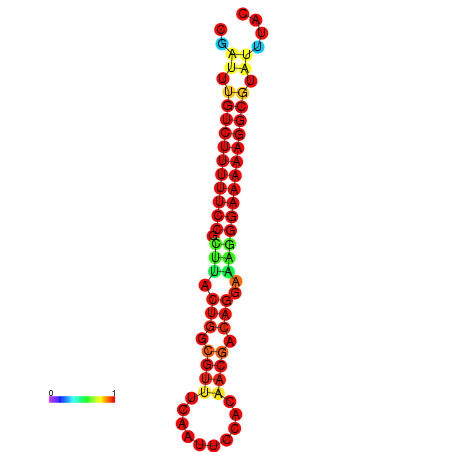
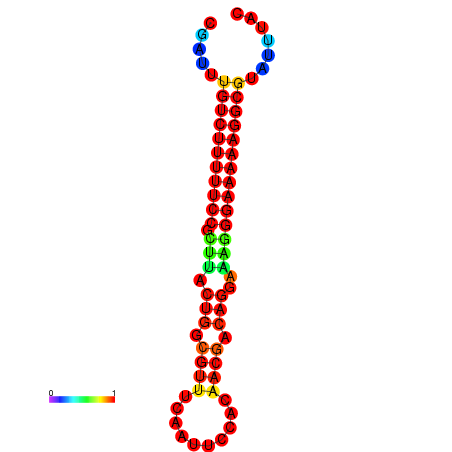
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:21661799-21661917 - | GAAAA--------------------------------------------GTGGATACAC-AAATTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTCAATTCCACAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAGTTTGCTCAGCGATT------------------------------TGCA------------------------------------ |
| droSim1 | chr3L:21004826-21004944 - | GAAAA--------------------------------------------GTGGATACAC-AAATTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTCATTTCCACAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAGTTTGCCCAGCGACT------------------------------TGCA------------------------------------ |
| droSec1 | super_11:1581867-1581985 - | GAAAA--------------------------------------------GTGGATACAC-AAATTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTCATTTCCACAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAGTTTGCCCAGCGACT------------------------------TGCA------------------------------------ |
| droYak2 | chr3L:18849495-18849613 + | GAAAA--------------------------------------------GTGGATACAC-AAATTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTCATTTCAACAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAGTTTGACCAGCGATT------------------------------TGCA------------------------------------ |
| droEre2 | scaffold_4784:21318808-21318933 - | GAAA-------------------------------------TACACAAAGTGGATACAC-AAATTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTCATTTCAACAACGACAGGAAAGGGAAAAAGGCGTATTCACTATGAGTTTGGCCAGCGACT------------------------------TGCA------------------------------------ |
| droAna3 | scaffold_13337:3985740-3985858 - | CAAGA--------------------------------------------ACATATATTC-AAATTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTAATTCAAACAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAGTTTGGTCAACATTT------------------------------CGTC------------------------------------ |
| dp4 | chrXR_group6:4548558-4548677 + | GTAAA----------------------------------A-TATACAAAG-----ATC-AAAATTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTTATTCGATCAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAGTTTATCCAACAA---------------------------------ACA------------------------------------ |
| droPer1 | super_9:2845596-2845715 + | GTAAA----------------------------------A-TATACAAAG-----ATC-AAAATTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTTATTCGATCAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAGTTTATCCAACAA---------------------------------ACA------------------------------------ |
| droWil1 | scaffold_180698:1092691-1092806 + | ATATA--------------------------------------------ATGTATGTAT-AAATTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTTATTTAACTAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAGTTTAATCAACAAAT------------------------------T--------------------------------------- |
| droVir3 | scaffold_13049:24266823-24266971 + | ATATC--------------------------------------------TTTGATCTATAAAGTTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTTATT-GAGCAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAATTTGTTCAACCAATCTGTTTAAACACCTATTCAATGAAAAGAAATGCT------------------------------------ |
| droMoj3 | scaffold_6680:18891183-18891347 + | GATCTA--ACTGAATCGATTTT--------------------------------TATATAAAGTTCTAGTCGATTTGTCTTTTTCCGCTTACTGACGTTTTATT-GAACAACGGCAGGGAAGGGAAAAAGGCGTATTTACTATGAATTATTTCAACGATA------------------------------TAAAAATTTCTATTAAAATAATAATCAGAGTGAGAGAGGG |
| droGri2 | scaffold_15110:9482044-9482193 - | TACCAGAGACTAAA-CATTTTGTGTATAAGTATTGTATGATTAAGCAATC-----TTC-AGAGTTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTTATT-GAGCAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAATGTAATCAACAA------------------------------------------------------------------------ |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:21661799-21661917 - | GAAAA--------------------------------------------GTGGATACAC-AAATTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTCAATTCCACAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAGTTTGCTCAGCGATT------------------------------TGCA------------------------------------ |
| droSim1 | chr3L:21004826-21004944 - | GAAAA--------------------------------------------GTGGATACAC-AAATTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTCATTTCCACAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAGTTTGCCCAGCGACT------------------------------TGCA------------------------------------ |
| droSec1 | super_11:1581867-1581985 - | GAAAA--------------------------------------------GTGGATACAC-AAATTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTCATTTCCACAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAGTTTGCCCAGCGACT------------------------------TGCA------------------------------------ |
| droYak2 | chr3L:18849495-18849613 + | GAAAA--------------------------------------------GTGGATACAC-AAATTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTCATTTCAACAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAGTTTGACCAGCGATT------------------------------TGCA------------------------------------ |
| droEre2 | scaffold_4784:21318808-21318933 - | GAAA-------------------------------------TACACAAAGTGGATACAC-AAATTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTCATTTCAACAACGACAGGAAAGGGAAAAAGGCGTATTCACTATGAGTTTGGCCAGCGACT------------------------------TGCA------------------------------------ |
| droAna3 | scaffold_13337:3985740-3985858 - | CAAGA--------------------------------------------ACATATATTC-AAATTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTAATTCAAACAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAGTTTGGTCAACATTT------------------------------CGTC------------------------------------ |
| dp4 | chrXR_group6:4548558-4548677 + | GTAAA----------------------------------A-TATACAAAG-----ATC-AAAATTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTTATTCGATCAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAGTTTATCCAACAA---------------------------------ACA------------------------------------ |
| droPer1 | super_9:2845596-2845715 + | GTAAA----------------------------------A-TATACAAAG-----ATC-AAAATTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTTATTCGATCAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAGTTTATCCAACAA---------------------------------ACA------------------------------------ |
| droWil1 | scaffold_180698:1092691-1092806 + | ATATA--------------------------------------------ATGTATGTAT-AAATTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTTATTTAACTAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAGTTTAATCAACAAAT------------------------------T--------------------------------------- |
| droVir3 | scaffold_13049:24266823-24266971 + | ATATC--------------------------------------------TTTGATCTATAAAGTTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTTATT-GAGCAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAATTTGTTCAACCAATCTGTTTAAACACCTATTCAATGAAAAGAAATGCT------------------------------------ |
| droMoj3 | scaffold_6680:18891183-18891347 + | GATCTA--ACTGAATCGATTTT--------------------------------TATATAAAGTTCTAGTCGATTTGTCTTTTTCCGCTTACTGACGTTTTATT-GAACAACGGCAGGGAAGGGAAAAAGGCGTATTTACTATGAATTATTTCAACGATA------------------------------TAAAAATTTCTATTAAAATAATAATCAGAGTGAGAGAGGG |
| droGri2 | scaffold_15110:9482044-9482193 - | TACCAGAGACTAAA-CATTTTGTGTATAAGTATTGTATGATTAAGCAATC-----TTC-AGAGTTCTAGTCGATTTGTCTTTTTCCGCTTACTGGCGTTTTATT-GAGCAACGACAGGAAAGGGAAAAAGGCGTATTTACTATGAATGTAATCAACAA------------------------------------------------------------------------ |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
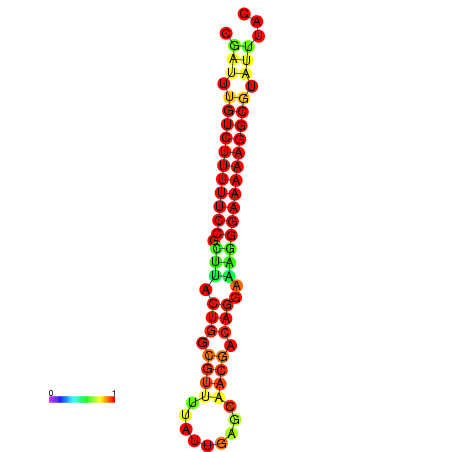
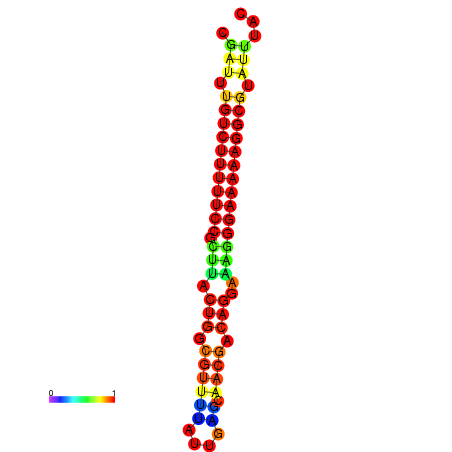
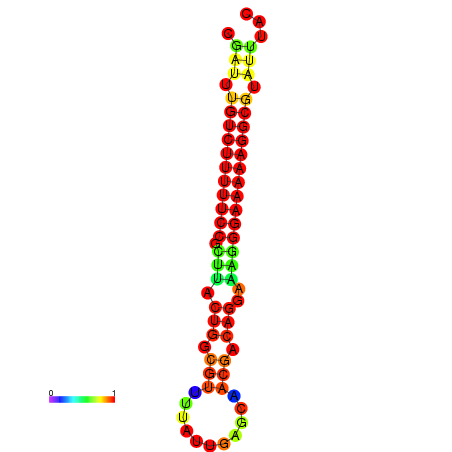
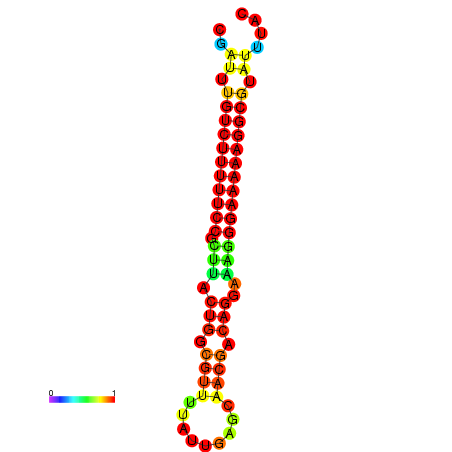
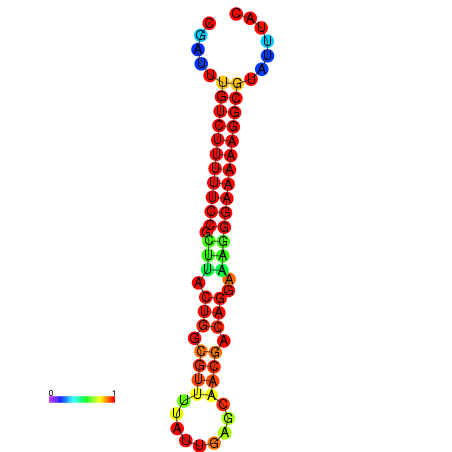
| dG=-21.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
 |
| dG=-21.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 |
|---|---|---|---|
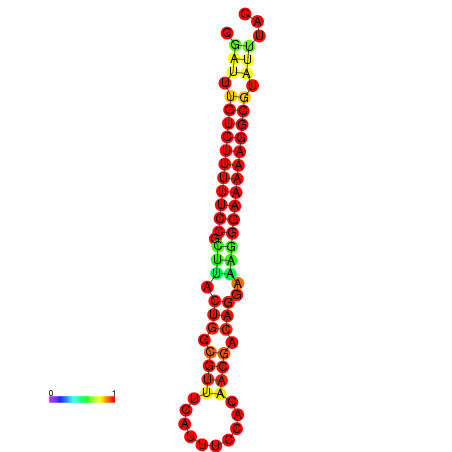 |
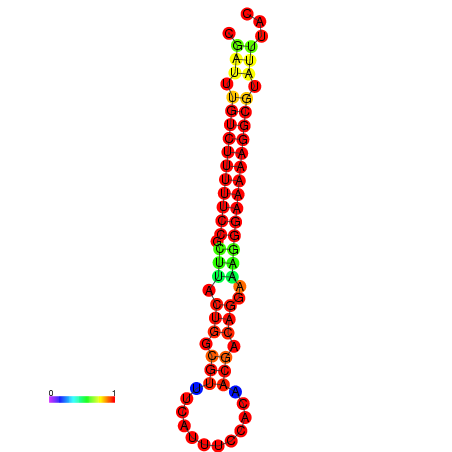 |
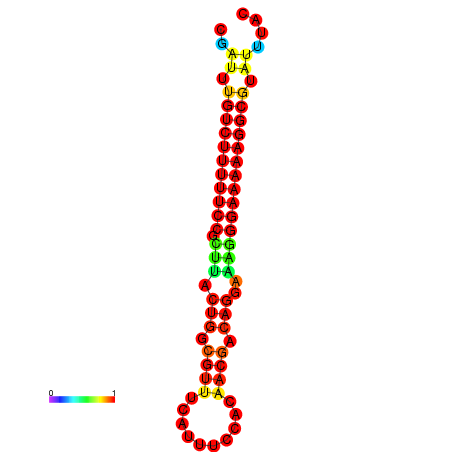 |
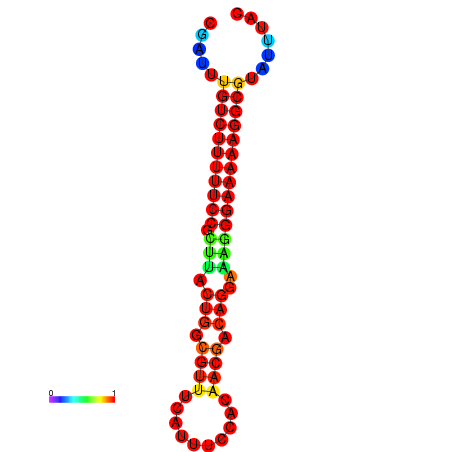 |
| dG=-21.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 |
|---|---|---|---|
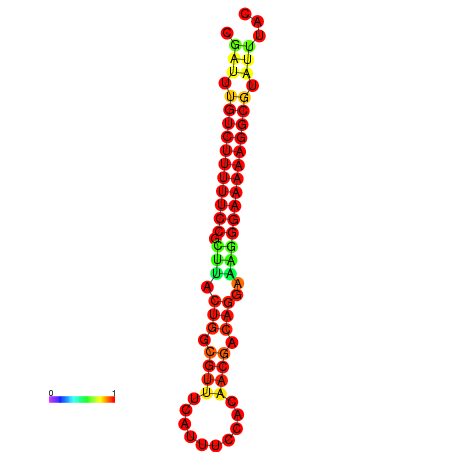 |
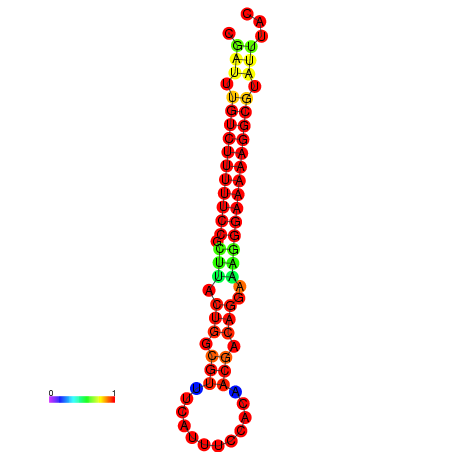 |
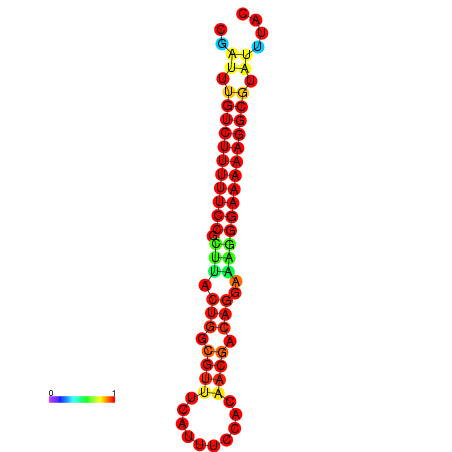 |
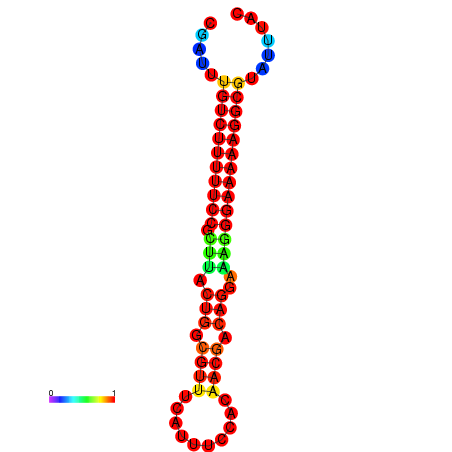 |
| dG=-21.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 |
|---|---|---|---|
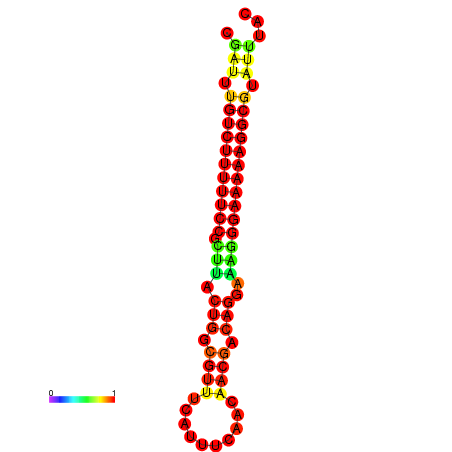 |
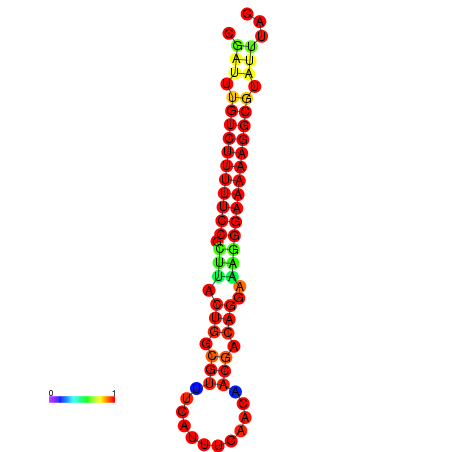 |
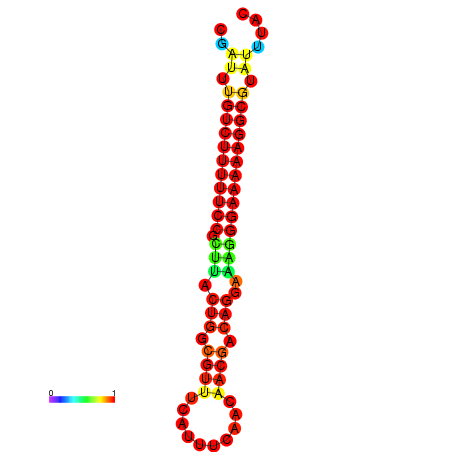 |
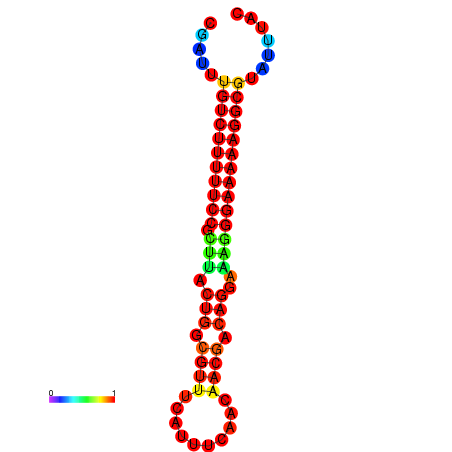 |
| dG=-21.2, p-value=0.009901 | dG=-20.3, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 |
|---|---|---|---|---|
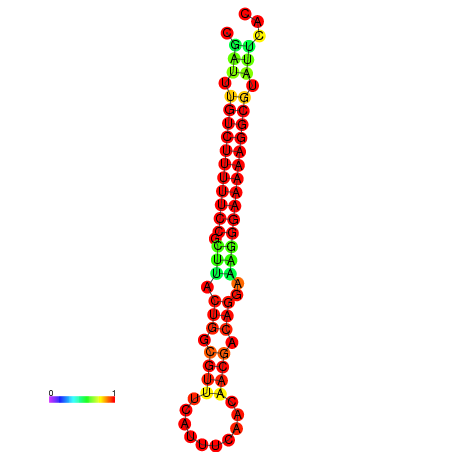 |
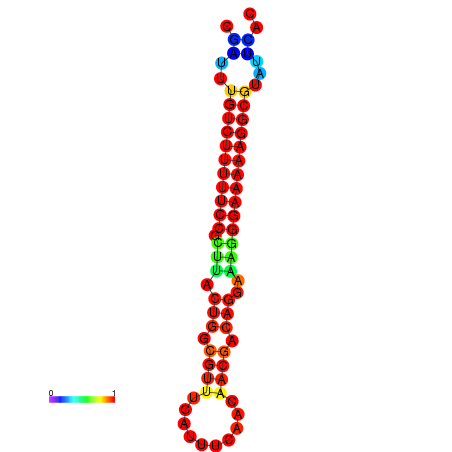 |
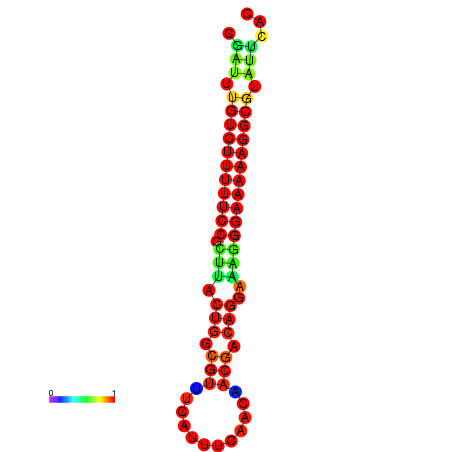 |
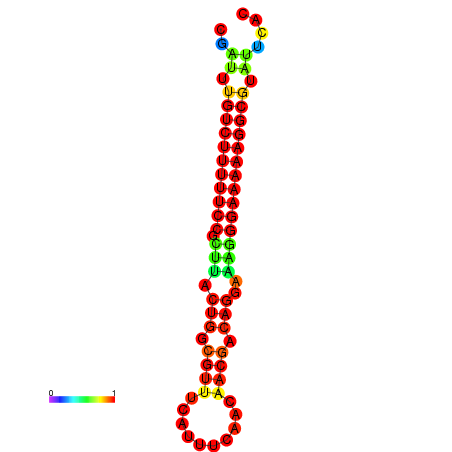 |
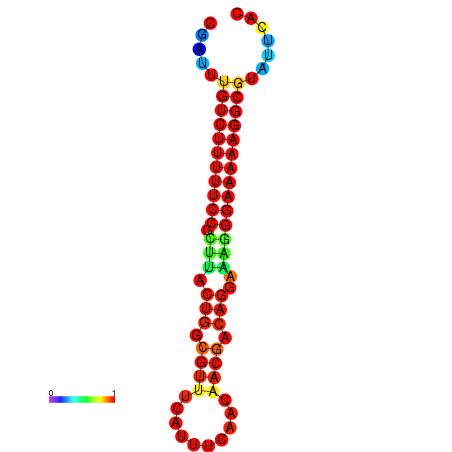 |
| dG=-21.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 |
|---|---|---|---|
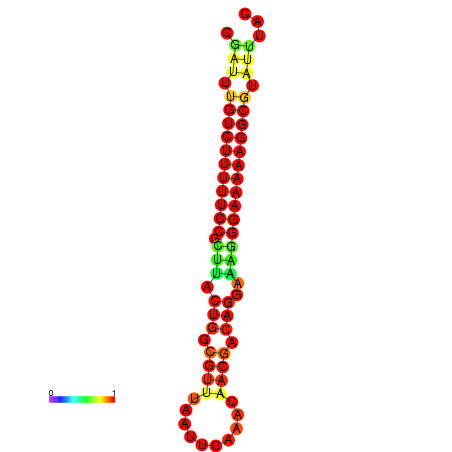 |
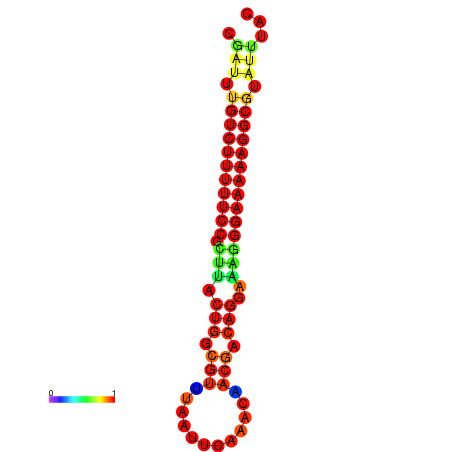 |
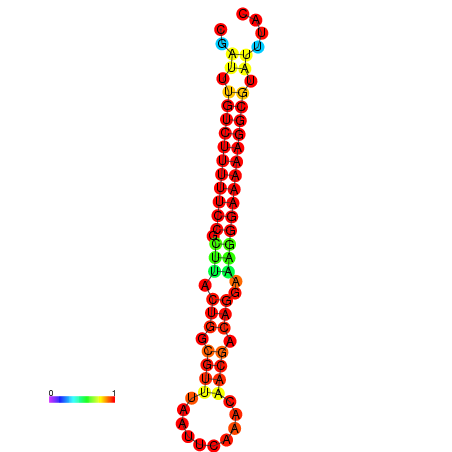 |
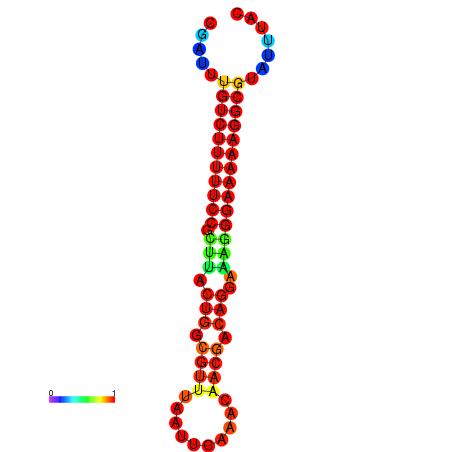 |
| dG=-21.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 |
|---|---|---|---|
 |
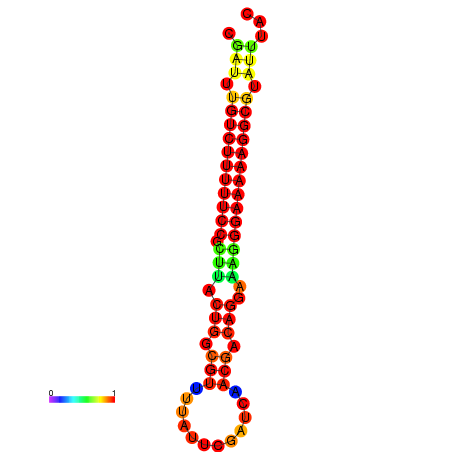 |
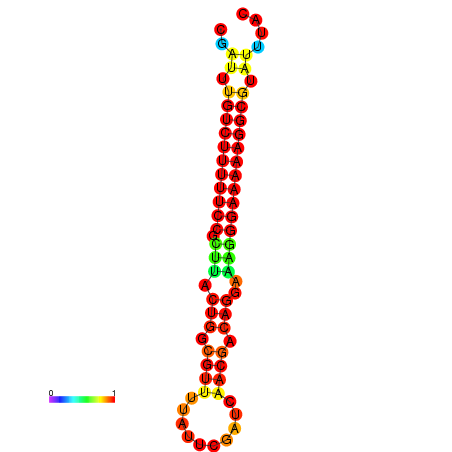 |
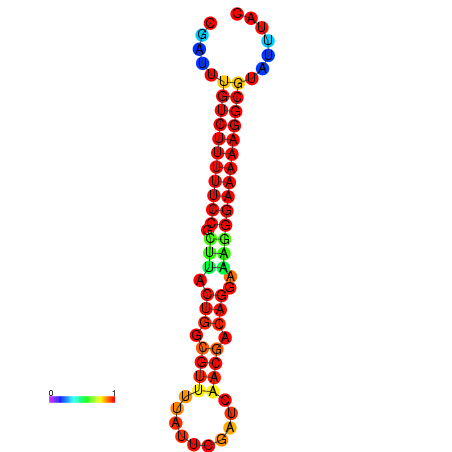 |
| dG=-21.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 |
|---|---|---|---|
 |
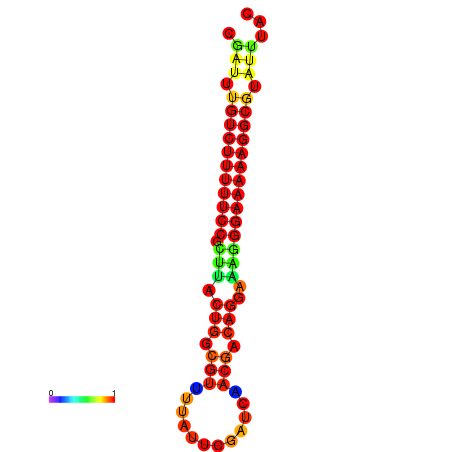 |
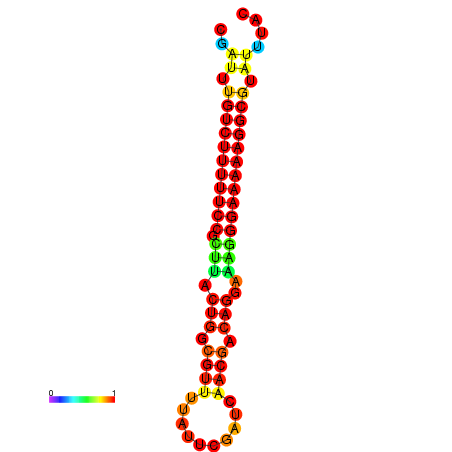 |
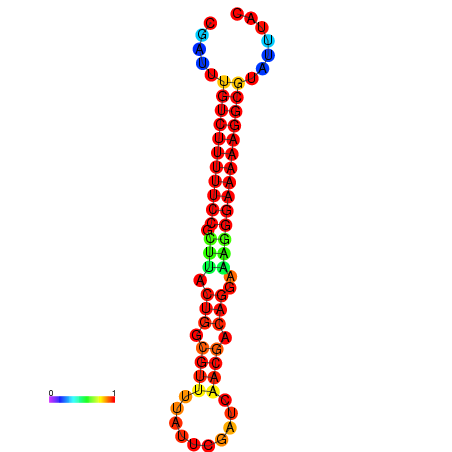 |
| dG=-22.0, p-value=0.009901 | dG=-21.0, p-value=0.009901 | dG=-21.0, p-value=0.009901 |
|---|---|---|
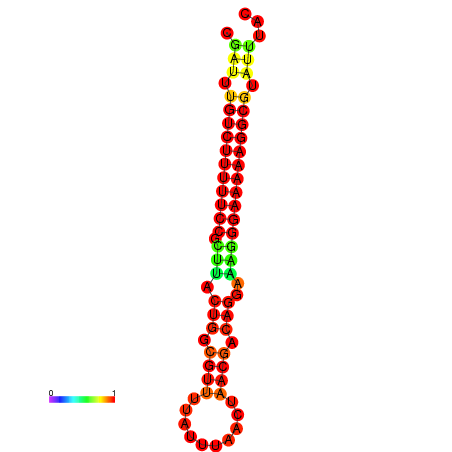 |
 |
 |
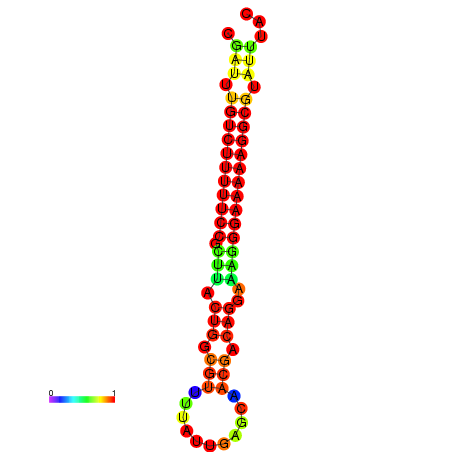
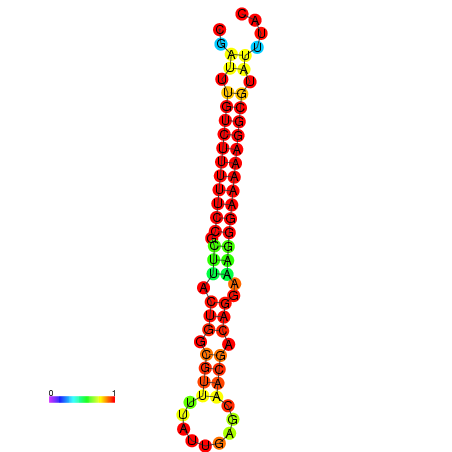
| dG=-21.3, p-value=0.009901 | dG=-20.6, p-value=0.009901 | dG=-20.3, p-value=0.009901 | dG=-20.3, p-value=0.009901 | dG=-20.3, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
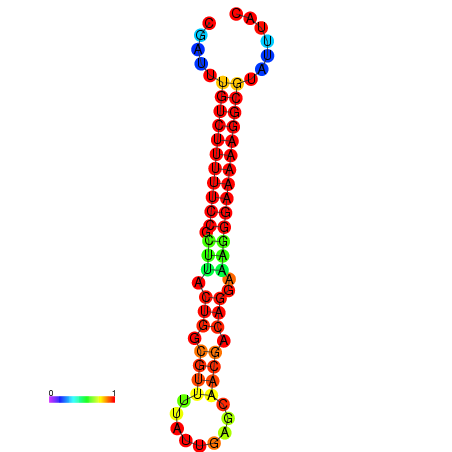 |
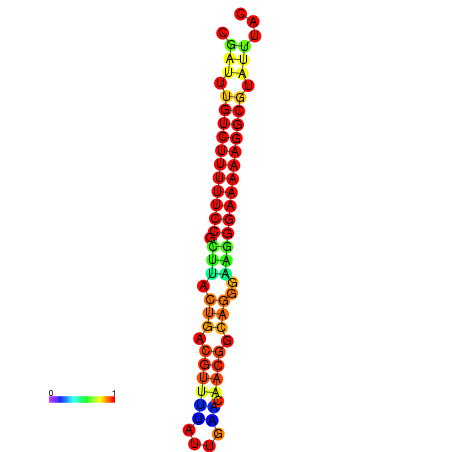
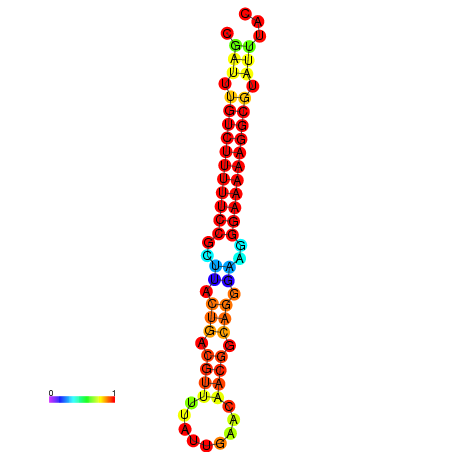
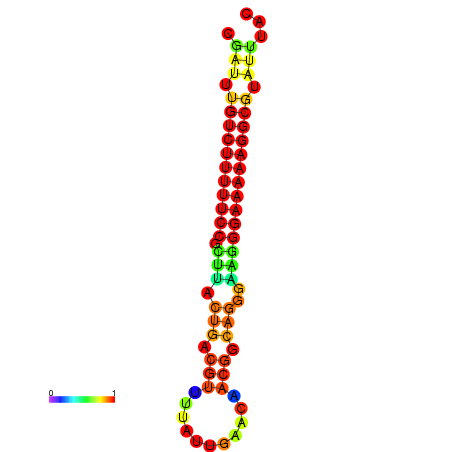
| dG=-21.3, p-value=0.009901 | dG=-20.5, p-value=0.009901 | dG=-20.5, p-value=0.009901 | dG=-20.3, p-value=0.009901 | dG=-20.3, p-value=0.009901 |
|---|---|---|---|---|
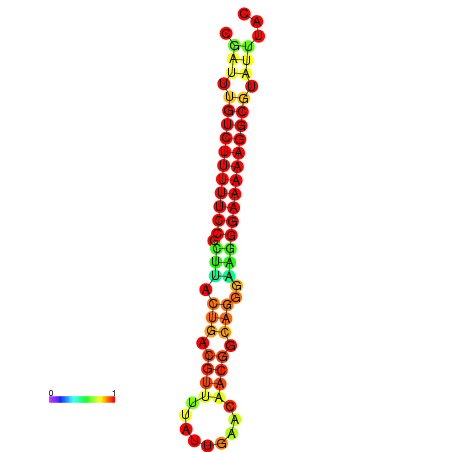 |
 |
 |
 |
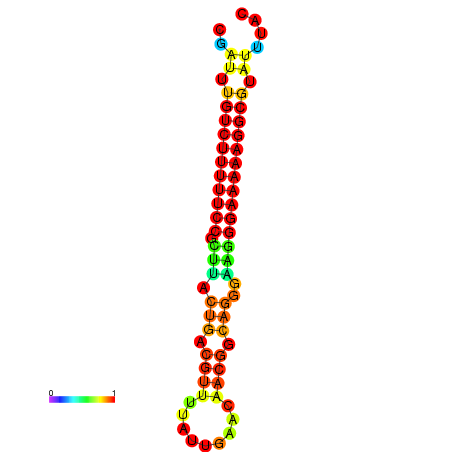 |
| dG=-21.3, p-value=0.009901 | dG=-20.6, p-value=0.009901 | dG=-20.3, p-value=0.009901 | dG=-20.3, p-value=0.009901 | dG=-20.3, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
Generated: 03/07/2013 at 05:19 PM