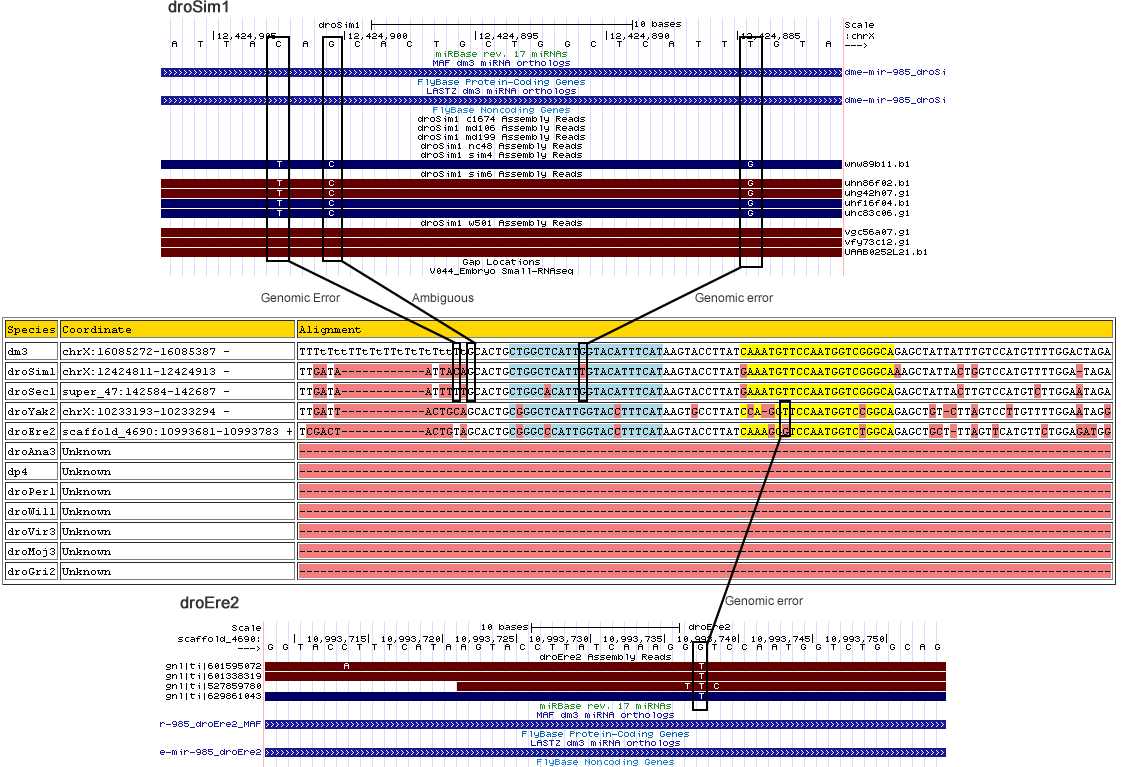
dme-mir-985
Changes
| Species | Chrom | Pos | Ref base | Type | A | C | G | T | RNAseq Total | A | C | G | T | Trace Total | Decision | Partition |
| droEre2 | scaffold_4690 | 10993738 | G | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | Genomic Error | miR |
| droSim1 | chrX | 12424885 | A | 3 | 0 | 0 | 0 | 0 | 0 | 0.375 | 0.625 | 0 | 0 | 8 | Genomic Error | miR* |
| droSim1 | chrX | 12424901 | C | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0.375 | 0.625 | 0 | 8 | Ambiguous - Suggested base change does not agree with neighboring species |
3' MOR |
| droSim1 | chrX | 12424903 | G | 3 | 0 | 0 | 0 | 0 | 0 | 0.625 | 0 | 0.375 | 0 | 8 | Genomic Error | 3' MOR |

Corrected alignment
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:16085272-16085387 - | TTTATAATTATATTATATATAATAGCACTGCTGGCTCATTGGTACATTTCATAAGTACCTTATCAAATGTTCCAATGGTCGGGCAGAGCTATTATTTGTCCATGTTTTGGACTAGA |
| droSim1 | chrX:12424811-12424913 - | TTGATAATTA------------TAGCACTGCTGGCTCATTGGTACATTTCATAAGTACCTTATGAAATGTTCCAATGGTCGGGCAAAGCTATTACTGGTCCATGTTTTGGA-TAGA |
| droSec1 | super_47:142584-142687 - | TTGATAATTT------------TTGCACTGCTGGCACATTGGTACATTTCATAAGTACCTTATGAAATGTTCCAATGGTCGGGCAGAGCTATTACTTGTCCATGTCTTGGAATAGA |
| droYak2 | chrX:10233193-10233294 - | TTGATTACTG------------CAGCACTGCGGGCTCATTGGTACCTTTCATAAGTGCCTTATCCA-GGTTCCAATGGTCCGGCAGAGCTGTC-TTAGTCCTTGTTTTGGAATAGG |
| droEre2 | scaffold_4690:10993681-10993783 + | TCGACTACTG------------TAGCACTGCGGGCCCATTGGTACCTTTCATAAGTACCTTATCAAAGGTTCCAATGGTCTGGCAGAGCTGCT-TTAGTTCATGTTCTGGAGATGG |
| droAna3 | Unknown | -------------------------------------------------------------------------------------------------------------------- |
| dp4 | Unknown | -------------------------------------------------------------------------------------------------------------------- |
| droPer1 | Unknown | -------------------------------------------------------------------------------------------------------------------- |
| droWil1 | Unknown | -------------------------------------------------------------------------------------------------------------------- |
| droVir3 | Unknown | -------------------------------------------------------------------------------------------------------------------- |
| droMoj3 | Unknown | -------------------------------------------------------------------------------------------------------------------- |
| droGri2 | Unknown | -------------------------------------------------------------------------------------------------------------------- |